Scripts
Take AcqKnowledge to the next level with BIOPAC Basic Scripting
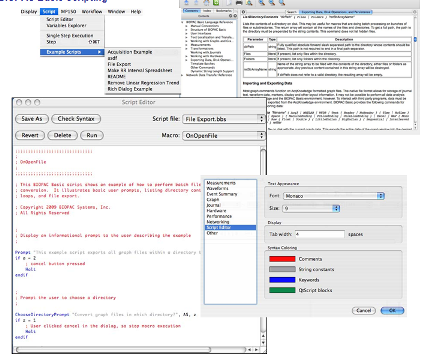 Scripts are easily created within AcqKnowledge and do not require programming experience, but a familiarity with scripting will certainly help. The commands are documented in the online reference library and example Script Downloads assist in the learning and development process.
Scripts are easily created within AcqKnowledge and do not require programming experience, but a familiarity with scripting will certainly help. The commands are documented in the online reference library and example Script Downloads assist in the learning and development process.
Significantly reduce analysis time and improve consistency with standardized automated procedures!
Customize the AcqKnowledge display—add customized messages and prompts to simplify a protocol, create a standardized procedure and include onscreen instructions, simplify the user interface to just the tools and menus used, etc.
Control AcqKnowledge functions—automate a calibration or analysis routine…BIOPAC Basic Scripting is used to create all of the AcqKnowledge analysis routines, which provide useful building blocks for creating further analysis routines.
Consolidate a series of complex tasks into one automated routine—perform batched analysis to eliminate repetitive tasks & eliminate the potential for error when running the same procedure many times across multiple subjects.
Improve the quality of data with greater standardization and consistency—guarantee reproducibility, no matter who is controlling the experiment!
 Add the power of BIOPAC Basic Scripting to your lab with a license for Windows or Mac, or try a Developer Bundle with Basic Scripting License.
Add the power of BIOPAC Basic Scripting to your lab with a license for Windows or Mac, or try a Developer Bundle with Basic Scripting License.
Script Downloads with Demo Screencasts
Script 000 – Output options: display warning, write to journal, exportScript 001 – Copy Markers
Script 002 – Place marker at index
Script 003 – Place marker at peak
Script 004 – Custom multiple measurements at marker
Script 005 – Mark TTL start and end
Script 006 – Mark TTL start and end with interval at end
Script 007 – Mark selection
Script 008 – Interpolate marked segments
Script 010 – Hide all channels
Script 011 – Show all channels
Script 015 – Average all waveforms
Script 016 – Average all visible waveforms
Script 017 – Automatic ensemble average all waveforms based on marker
Script 020 – fNIR Set channel labels
Script 021 – fNIR Show Oxy channels
Script 022 – fNIR Show Deoxy channels
Script 023 – fNIR Show left hemisphere Oxy channels
Script 024 – fNIR Show left hemisphere Deoxy channels
Script 025 – fNIR Show right hemisphere Oxy channels
Script 026 – fNIR Show right hemisphere Deoxy channels
Script 027 – Delete selected channel
Script 028 – Prepare ECG and place QRS markers
Script 029 – Keep QRS markers only
Script 030 – Get Heart Rate from QRS markers
Script 031 – Export Mean HR in Excel
Script 036 – Batch process
Script 039 – Import Markers from Text
Script 040 – Convert Numerical Marker Labels to Text
Script 046 – Analog Input to Stim Events
Script 047 – Import Markers from Noldus ObserverXT
Script 048 – Stop Data Acquisition by Trigger
Script 052 – Metabolic Analysis
Script 053 – Convert variable length TTL to unique markers
Script 054 – Ensemble Average Segments
Script 055 – Analyze RSA Using HRV (Segmented HRV)
Script 056 – Compute HRV Statistics
Script 057 – Displaying AcqKnowledge Data in Vizard as Text in Real Time using NDT
Script 058 – Biofeedback and VR
Script 059 – Sending Markers from Vizard to COBI
Script 060 – Dynamometry Feedback (MVC line trace)
Script 061 – Offset the last marker by a set time period
Script Tool – ID Codes for Event Markers
Scripting – Feedback Control
Scripts to Prepare ECG data for HRV Analysis
Stay Connected