EDA Data Analysis & Correction
EDA Questions and Answers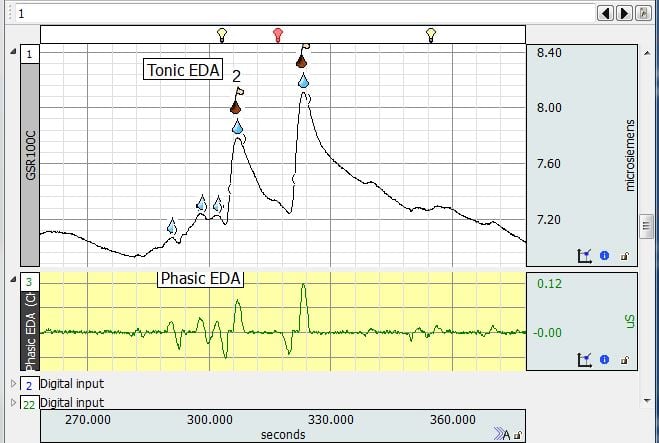
Your colleagues submitted these questions during BIOPAC EDA Webinars.
Topics include
- EDA Artifact Detection & Correction
- EDA Data Measurement & Analysis
Other Sections include
If your question isn’t covered, learn more in EDA: Electrodermal Activity Applications, review EDA Signals & Measurements, or contact BIOPAC Support.
EDA Data Measurement & Analysis
- Q: How do I preprocess and analyze my data sets?A: I would recommend watching some of our earlier webinars because they cover recording and preparing the data for analysis. The following link will take you to the webinar section of the website. https://www.biopac.com/webinars/
- Q: What is EDA signal? How it is being recorded? Why do we need to record EDA instead of recording EMG? What is the relation between SCR, EDA, GSR? What is the difference between EMG & EDA?
A: A good discussion and differentiation of these signals can be found in Handbook of Psychophysiology. John T. Cacioppo, Gary Berntson, Louis G. Tassinary and Psychophysiology: Human Behavior and Physiological Response by John L. Andreassi.
A: The webinar covers everything from subject, hardware and software setup, to recording great data and analyzing the signals.
- Q: How should we automate preparation (e.g., cleaning) of EDA data, checking data quality, and filtering?
A: The presentation provides a good overview of the recommended steps for preparation of data and examples of good vs. bad data. The recorded EDA Analysis and Scripting Webinar shows how these functions can be automated using BIOPAC scripting. These functions will also be addressed in the EDA Analysis Essentials Webinar. Finally, we recommend you consult the EDA resources in our Knowledge Base and Application Notes.
- Q: GSR peaks indicate emotional arousal. They are identified by eyeball inspection. How can such peak identification be automated?
A: The webinar demonstrates two automated techniques for identifying and scoring Skin Conductance Responses. AcqKnowledge includes fully-automated routines for EDA analysis.
- Q: Different ways of analyzing EDA data (tonic vs. phasic responses)
A: The presentation shows how to acquire both the Tonic and Phasic signals and walks through a variety of analysis options.
- Q: Do you have any tips for using AcqKnowledge to compare background tonic SCL across multiple periods within a file?
A: The Focus Areas are a great tool for comparing segments of data. I demonstrated the use of the Focus Areas in the webinar and they were also covered in the previous webinar. We also have a screencast that provides additional information. https://www.biopac.com/events/acqknowledge-webinar/ , https://www.youtube.com/watch?v=CBYGYklB9Yo
- Q: We record continuously remotely we want to segment periods of interest and compare reactivity scoring within and across controlsA: If you want to manually identify the periods of data, you can use the Focus Areas to highlight the regions you want to analyze. If you use the SCR analysis routine, it will score the data and you can then use the Epoch analysis to take measurements from the Focus Areas.
- Q: How to batch mode analysis in AcqKnowledge?
A: The following link will take you to a webinar that covers analysis and scripting. The BIOPAC basic scripting tool allows you to batch process analysis and automate much of the analysis that were included in this webinar: https://www.biopac.com/events/eda-webinar-analysis-scripting/
- Q: What would be the best measure to report for tonic EDA? Frequency, Amplitude or SD of NS-SCR?
A: We refrain from advising on the best measure. We recommend that you consult the reference literature that was included in the presentation as well as the literature on the topic of EDA in general. The analysis routines in AcqKnowledge will provide you with a wide range of options to choose from and all of the frequently used and recommended measures.
- Q: Does the Connect Endpoint function use a linear model to connect the points?
A: It draws a line from the first selected sample point to the last selected sample point and interpolates the values on this line to replace the original data.
- Q: Can I just study using the raw EDA data? Or should I edit the data (e.g., smoothing?) beforehand?
A: If the signal is clean, it can be used as is. But you can resample to 50 Hz (no less than 50 Hz) in order to speed up the performance of the analysis algorithms.
- Q: Why was median smoothing selected? How was the 50 setting selected?
A: Median smoothing rejects outliers while mean smoothing averages them into the result. It will eliminate rap transient spikes from a slow moving signal such as electrodermal activity. 50 samples were selected because the sample rate was 50 Hz. Usually, a smoothing window equal to the number of samples per second will remove most artifacts while not disturbing the physiological trends in the data. Remember to apply a 1 Hz FIR LP afterwards, to smooth out the result. You can perform testing with these and other settings on clean data and compare to the original to see that the data are not significantly altered by the transformations. We recommend such testing steps before transforming data in general. See also Methods for Computing Phasic EDA.
- Q: Will using smoothing on a signal remove some information that might otherwise be useful, such as skin conductance responses or altering the SCL?
A: No, median smoothing will not impact the signal if it is applied correctly and it is easy to determine whether you have altered the signal by overlapping the raw and smoothed waveforms. If the filter becomes too aggressive (if you use too many samples for the window), then it will also transform the underlying trends in the signal. For data sampled at N samples per second, a median smoothing filter with window size N will result in a slight reduction of observed P-P changes in responses:
You have to define the limits for an acceptable transformation. In this example, the peak-to-peak measurement after smoothing and low-pass filtering at 1 Hz differs by about 3% from the raw data (0.337 vs 0.347). But a filter this aggressive will eliminate most fast motion artifacts. If this trade-off is acceptable it’s the researcher’s decision and we recommend to always perform testing on both clean and noisy data before choosing a strategy to remove artifacts.
- Q: You selected “Smoothing Baseline Removal.” If I want to compare the data of baseline (before sound stimuli) and data of sound exposure, should I still choose that?
A: Smoothing baseline removal was used during the webinar as a method to obtain the phasic EDA signal, the signal that represents changes in EDA. If you want to compare the participant’s responses during two different blocks of the experiment (such as when sound stimuli are presented vs baseline), please refer to the section of the webinar that covered the block analysis.
- Q: Can you look at batches of waveforms from different subjects in addition to batch analysis?
A: Yes.
- Q: Are there any sources you can provide for how to run statistical tests using the data obtained from BIOPAC?
A: We would like to refrain from making recommendations on how to run statistical tests.
- Q: Can the analysis (as shown at this moment) be applied to multiple participants without having to do it manually for each participant?
A: Yes, and this will be covered in the follow-up webinar.
- Q: Why resample? Is there a problem with having it at the 2,000 Hz rate of data collection?
A: Reducing the sample rate lessens the computational load for the analysis.
- Q: Is it possible to get access to graphs/pictures of what the EDA signal will look like when different issues arise (e.g., how does EDA look when electrodes have expired, when the wrong gel is used, when the participants are moving a lot, when there is momentary decoupling, etc.).
A: A separate troubleshooting guide is being prepared and it will contain actual data, not just pictures. We should focus on good data—show what the signal should look like and not worry about what bad data looks like. I do not like this approach…there are just too many potential issues.
- Q: We find that while we are able to get a good EDA signal initially, the signal turns into noise within 20 minutes of the protocol. Can you offer any insights?
A: It is best to send over a data sample to support@biopac.com. We would need to see the data to provide useful advice. Please send the raw .acq file. However, I would start by looking at the electrode to subject connection and make sure that everything is good there. Check the quality of the electrodes and ensure they are making good contact with the subject. When you send a file make sure that you include a full description of your equipment and participant setup including any tasks the subject is performing.
- Q: How do you analyze SCR that has both anticipatory and stimulus dependent responses within a short time window?
A: The AcqKnowledge cycle detector can take measurements around specific stimulus events, both before the event and after the event, to automate the extraction of measurements. If you use the automated Event-related EDA Analysis option, the software will identify specific and nonspecific skin conductance responses within the recording. You can then use the Find cycle detector to measure SCL and count SCRs during the anticipatory phase of the recording. We have several Find Cycle screencasts that demonstrate how to use the Find Cycle detector. I would also refer to the guidelines to see what is recommended for measurement timing intervals.
- Q: How would you go about analyzing unspecific EDA data?
A: This will be similar to the Block demonstration I gave in the presentation, but instead of blocks, you can look at responses over defined time intervals – e.g., every 2 minutes. AcqKnowledge has a fully automated Epoch analysis routine that will allow you to automatically measure the data and export the results to Excel. The Epoch Analysis screencast will provide some additional information about this feature.
- Q: I notice a lot of variability among participants’ EDA data, in spite of them all being subjected to the same experimental manipulation. This variability makes it difficult to test the hypotheses even when the sample size is good (e.g., 30-40 participants per group in a 2- or 3-group between-groups design). I am already following the same protocol with all participants; is there anything else I can do to reduce this variability? I observe this outcome when using either MP100 or MP45 system.
A: I’m not clear what you mean by EDA variability because there are two primary signals – Tonic and Phasic. The Skin Conductance Level will vary quite a lot among participants because this is the absolute signal, the tonic waveform. However, the Skin Conductance Response analysis determines how responsive a subject is and also measures the size and amplitude of each response. The software measures the SCR amplitude by measuring SCL at the point of SCR onset, and again at the peak of the SCR, and then subtracting the onset value from the peak value to provide the amplitude value. The size of the response is relative to the SCL at the point the response started. If you are concerned about the quality of your data, you can contact our Support Department and they will gladly provide you with some specific feedback.
- Q: How to standardize and compare one’s EDA data with others?
A: If you are trying to compare data that has been previously published, you can, in most cases, adjust the settings in AcqKnowledge‘s automated analysis routines to match those described in the publication. I would also recommend consulting the list of references that are included in the webinar slide deck to determine current recommendations for EDA analysis. BIOPAC has created an automated solution that conforms to the guidelines but also provides users with flexibility to match other analysis strategies.
- Q: What is the value of viewing the EDA response with a ball pushing a bar rather than viewing it as a graph/in the format it is being recorded?
A: This example was a part of a biofeedback game that uses EDA and AcqKnowledge‘s Network Data Transfer functionality to provide real-time access to the data for biofeedback. A sphere would materialize after every SCR (skin conductance response). If the goal is for the participant to relax, they try to keep their responses down or else the bar will be tipped over. In another version, the game can be played against a human or computer with the goal of generating more and larger responses than the opponent in order to knock down the bar first. The game adapts the size of the spheres to the max response of the participant so far, thus accounting for individual variability.
- Q: Is the adaptive scaling available in Biopac Student Lab software?
A: Yes.
- Q: Do you use absolute or relative parameters for data error/rejection?A: If you are reviewing the data before the analysis is performed then data quality is a little subjective. During this part of the review, you should be looking at the absolute Skin Conductance Level data, which is the Tonic waveform and make sure that there are no obvious sections of artifact such as sudden jumps or drops in the data. However, the analysis uses specific parameters for rejecting data such as the threshold crossing and the ability to reject any SCR below X% of the maximum SCR response. The default setting for this rejection is 10%, but you can set the value to zero and every SCR will be included in the analysis.
- Q: which is the correct sampling rate for biomedical signals?A: Each physiological signal has its own sample rate which is based on the frequency components within the signal. EDA is a slow signal so you can reliably record the data at 70-100 samples a second to correctly capture the signal. However, if you are looking at facial EMG, the frequency components within the EMG waveform are much higher and you would want to sample that data at >1,000 samples a second.
- Q: What do you use for a calibration signal?A: If you use the module setup function in AcqKnowledge, the system will scale the data for you and you will not require a calibration signal. The system will automatically guide you through this process.
- Q: Is it possible to do deconvolution analysis with AcqKnowledge? What is your opinion of deconvolution-based approaches to measuring SCRs?
A: AcqKnowledge does not currently include an automated deconvolution analysis option, but we are constantly reviewing feature requests. AcqKnowledge can export the data in 3rd-party formats to aid in the process.
EDA Artifact Detection & Correction
- Q: Can you elaborate on how to identify electrical noise in the recording? As well as how to fix it when the participant is there or during the data cleaning stage?
A: Electrical noise is 50 Hz or 60 Hz. You can use the Spectrum analyzer palette (Display >Show >Spectrum analyzer palette) in real-time as well as after the recording. A peak will be obvious at 50 Hz/60 Hz (depending on your region). This is an unusual problem to have with the EDA data as it is low-pass filtered at the amplifier. This means the source of noise is not related to the EDA, with the most likely culprit being third party equipment that is connected to our system without optical isolation. Dry electrodes or poor contact between the electrode and the subject will also exasperate this problem.
- Q: How do I remove TTL artifact from GSR data prior to copying graph data?A: If you have the TTL triggers going to the digital channels on the system, you should not have artifacts on the raw data. Are you sure the artifacts are coming from the TTL triggers? If you review the earlier EDA webinar, we cover the use of the AcqKnowledge’ s, median smoothing function. This tool should allow you to remove such artifact without impacting the raw data. The median smoothing tool is a wonderful way to eliminate random spikes from slow moving data such as EDA. The following link will take you to the webinar: https://www.biopac.com/events/eda-webinar-part-1-recording-great-data/
- Q: You said fast SCL rise equals good data, is “fast drop of SCL” can be seen as a bad data (artifact)?
A: What’s important is to be aware that a SCR reaches the peak over 1-3 seconds while 50% decay may take anywhere from 2-10 seconds. So skin conductance data rises much faster than it drops. For instance, a very fast drop, for example a full 1 µS over less than 2 seconds, is unlikely to be physiological in nature.
- Q: For artifact correction: Should median smoothing be done before low-pass-filter or the other way around?
A: The median smoothing should be performed first to remove any high frequency artifacts from the signal before low pass filtering it. Low pass filtering will smooth artifacts into the data, so they should be removed first.
- Q: Where can I find some instructions on how to automate (i.e., generate a script) artifact correction?
A: Scripting is covered in this EDA Analysis & Scripting webinar.
- Q: Do you have any suggestions for removing artifact from mobile subjects?
A: It’s best to avoid artifact in the first place by using fresh electrodes and taping over the leads and electrodes. Experiment with placing electrodes in alternate locations that will not be influenced by movement that much but explore the literature beforehand. We have included one such reference in the presentation. If you are already done with the experiment, please refer to the answer of question 11. We are also working on new strategies for mobile applications that will help to prevent or minimize artifacts created during mobile recordings.
- Q: Please explain the automated artifact removal scripting in detail or provide additional information for this feature! Thanks!
A: This is discussed in detail during the follow-up webinar.
- Q: How do I recognize and remove artifacts created by movement?
A: These are changes in skin conductance that do not make sense given physiological expectations. Typically this means the EDA goes up or down at a very fast rate. Such artifacts are very fast. Here is an example:
Next, we have resampled the data to 50 Hz and applied a 50-sample median smoothing filter (Transform->Smoothing) and then 1 Hz FIR low pass filter (Transform > Digital filters > FIR filter > Low pass). We have zoomed in and the original data is seen on top with the transformed result on the bottom. The artifacts are completely eliminated:
This is even better illustrated by showing the waveform in scope mode, with the green waveform representing the clean result:
This is an example of the most typical type of artifact and method of correction. However, we will release an EDA troubleshooting guide in the future that will include AcqKnowledge sample files of various types of issues so you can learn how to handle the data yourself.
The follow-up webinar will address artifact removal automation techniques.
- Q: How do we quickly diagnose and fix low EDA signals: What to do when reading starts nice and smooth and then goes fuzzy? What causes the interference or artifacts that look like rectangular drops in the signal, and what is the best way to avoid/remove them? I noticed this in the data sample “untitled 3” that you had open at the very beginning. There is a lot of noise in my signal (e.g., the line is “hairy”). Is this something that will impact my data analysis?
A: The most common causes of low signal or poor data are poor electrode placement (location and/or adhesion) or the subject banging or knocking the electrodes. The subject banging or knocking the electrodes can also cause artifacts that are clearly not EDA. The presentation, along with BIOPAC Application Notes and Knowledge Base provides references on how to prevent issues from electrode placement. Use of a low-pass filter set to 1 Hz, as noted in the presentation, can help address some of the issues with “fuzzy” or noisy data. Artifact identification and removal (including motion artifacts), will be covered in the EDA Analysis Essentials Webinar and is covered in the recorded EDA Analysis and Scripting Webinar.
- Q: Can one use ICA to remove artifacts?
A: There are many techniques that can be employed to remove artifacts but we try and avoid spending too much time removing them by placing the electrodes and leads in locations that are less problematic. The tools in the webinar will provide you with some good examples for rapid data cleanup.
- Q: Respiration artifact correction! How to decipher a real SCR from a resp-related SCR?
A: When we test the EDA signal, we ask the participant to take a sharp, deep breath and to hold it for a second. This process will result in a response from most subjects. However, it is unlikely that a subject will perform this maneuver under normal circumstances. Normal breathing patterns will not result in a response but you can always record the respiration signal and check to see whether participants have irregular breathing patterns that may be influencing the data.
- Q: Is there a way to salvage data that is messy because of substantial movement/messing with the electrodes? For example, in our study on SCR among anxious participants our most anxious participants were the ones who played with the electrodes the most making their data (which is of most interest) also the messiest.
A: There is a lot that can be done to recover the data. You can use the median smoothing, connect endpoints and filtering techniques and/or contact us at support@biopac.com so we can help with further suggestions. We have seen literally thousands of files with EDA and can usually help to extract some useable data. However, it makes sense for you to consider using alternative electrode locations where participants are less likely to play with the electrodes and leads.
Stay Connected