Computing heart rate
Heart rate can be derived from various data sources but is not a physically measurable quantity, thus it is “computed” rather than measured.
A heart beat may be detected through a variety of measurements. Most commonly it is detected via ECG—measurements of the changes in the voltage across the chest. It may also be detected with a surface microphone, a photoplethysmogram (PPG) sensor, flow sensor, or through blood pressure measurements. Physiologically these different measurements are not all identical. ECG spikes indicate when the electrical events leading to a contraction of the heart muscle occur. A microphone indicates the timing of valve openings and closures, and these events are precipitated by the tensing and relaxing of cardiac muscle. PPG measures blood penetration into (typically) distal capillary beds. When the heart pumps, it increases the pressure locally and causes a pressure wave to travel through the circulatory system. Relative to the time of a heart beat as indicated by an ECG spike, the timing of capillary ingress or of a pressure wave and concomitant increase in blood velocity depend on where in the body the pressure or flow measurement is made. It is not necessarily true that the delay between these events is fixed even within a single recording session on a single subject.
All that said, heart beats or their after-effects are identified as some regular occurrence—typically of a positive peak—in the data. There are two main ways that rate information is extracted from such peaks using BIOPAC software.
- “Find Rate” – available as a calculation channel during data acquisition and via the “Analysis” menu (or “Transform” menu in older software versions) after data have been acquired. The algorithm used to derive rates via “Find Rate” or “Rate” calculation channels is extensively documented in Application Note 142.
- “Find Cycle” – available via the “Analysis” menu. Used in conjunction with the measurement boxes, Find Cycle provides some flexibility that Find Rate does not. In particular, Find Cycle may be used to insert event marks at positive peaks (as “qrs” events were placed in the file in the image above). These event marks can then be edited so that noise that initially causes erroneous “beats” to be identified or legitimate beats to be missed will not compromise the computation of the heart rate. Watch Video Tutorial: ECG Analysis using Find Cycle, which includes various editing tricks intended to make the process of cleaning the computational results more effective and efficient.
It is also possible to use “Find Cycle” to compute rates based on event marks placed by other algorithms. Many of the analyses built into AcqKnowledge actually use “Find Cycle” to insert event marks based on simple peaks in the data. On the other hand, “Detect and Classify Heartbeats” and “Locate [Human] ECG Complex Boundaries,” both available via the “Analysis” menu, use the OSEA beat detector algorithm and ecgpuwave respectively to identify and then event mark features within ECG data. After using one of these tools to place event marks, Find Cycle in conjunction with a BPM measurement can be used to find pairs of events indicating two successive beats. Thereby heart rate is computed cycle by cycle. With the BPM measurement saved as a channel in the graph, the heart rate is available for further computations (e.g., mean heart rate over long segments of data).
It is important to bear in mind that the “BPM” measurement is only intended to provide a measure based on exactly one inter-beat interval; the measurement is only meaningful when one and only one inter-beat interval is selected:
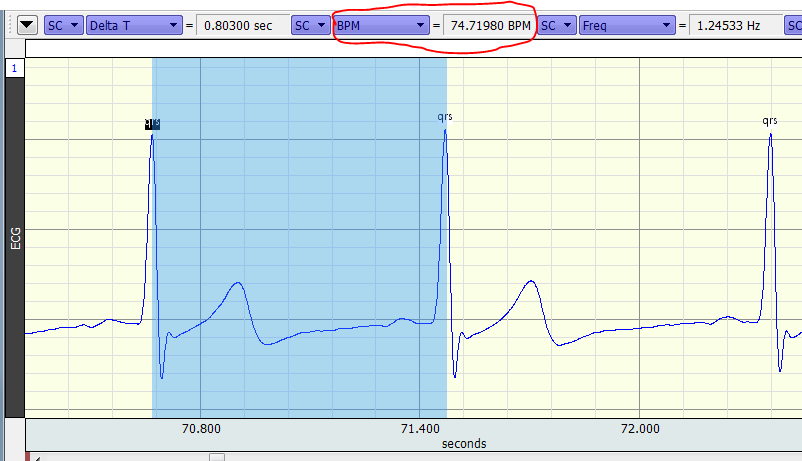
The measurement is not based on any numbers in the data file, only on the width (in time) of the area selected. The “BPM” measurement is useful for spot-checking (as above) or in conjunction with “Find Cycle” (see 2 above). If you highlight a segment of data containing multiple heartbeats, the BPM measurement provides a meaningless answer, NOT the average heart rate over the selected segment. To obtain an average heart rate, use a “Mean” measurement on a channel containing a computed rate.
last modified 19Jan2015
Stay Connected